Research
Overview
Research in the McBride lab revolves around three major themes in B cell biology: 1) How B cells in the tumor microenvironment affect patient immunotherapy response; 2) How DNA damage and repair contribute to immunoglobulin/antibody diversity; and 3) Interactions between B cells and oncogenic viruses and how they influence host-pathogen interactions. The lab undertakes a number of biochemical, molecular, and cellular approaches, while also using human tissue samples, mouse models, and a proprietary, McBride Lab-developed, high-throughput, single-cell immunoglobulin (Ig) cloning and antibody characterization pipeline. The pipeline is used for defining the immunoglobulin repertoire (the diverse range of B cell receptors expressed by an individual), and for identifying and selecting specific antigen-binding antibodies for recombinant production. This unique pipeline is key to many of the lab’s projects and collaborations.
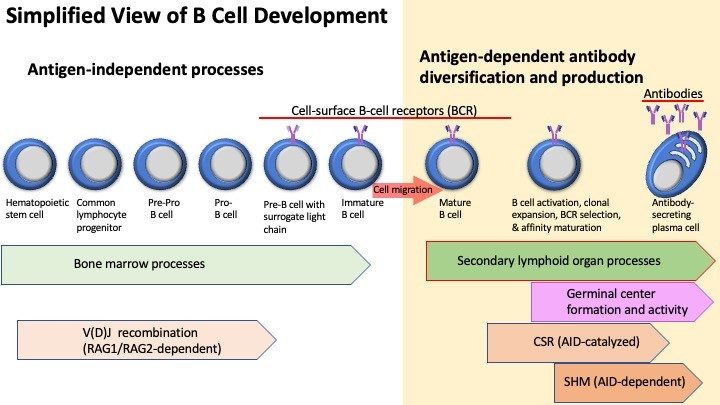
In mammals, B cells arise from hematopoietic stem cells in the bone marrow where they differentiate through multiple antigen-independent steps. The resulting naïve, immature B cell migrates to a secondary lymphoid organ, such as the spleen. There, antigen-induced B cell activation is coupled with gene expression changes in the cells of the microenvironment to give rise to the germinal center (GC).
Downstream events in antibody diversification depend on class-switch
DNA recombination (CSR) and somatic hypermutation (SMH). Both CSR and
SMH are influenced by activation-induced cytosine deaminase (AID).
During immune responses, mature B cells diversify immunoglobulin (Ig)
genes through SHM and CSR. SHM alters antibody affinity by introducing
nucleotide changes in the antigen-binding variable region of
antibodies. B cells producing B cell receptors with improved antigen
affinity are positively selected during affinity maturation and before
differentiating into antibody-secreting plasma cells.
B Cells in the Tumor Microenvironment and Patient Immunotherapy Response
The McBride group is breaking new ground in understanding B cell responses in tumor immunology. They are highly focused on comparing the immunoglobulin repertoire from patients who do and do not respond to immune checkpoint blockade (ICB), but they also have many other collaborative projects based on defining the immunoglobulin repertoire in different disease states and experimental conditions.
Tertiary lymphoid structures (TLSs) are lymphoid organs that form in non-lymphoid tissues, often in response to chronic inflammation. In tumors, the presence of TLSs correlates with better overall patient survival and better patient response to immunotherapy. Preliminary evidence from the McBride lab indicates that patients with a positive response to ICB may have B-cells that produce tumor-specific antibodies in the TLS. However, it is unclear how TLSs interact with the tumor microenvironment or how a specific tumor microenvironment impacts B cell function in the TLS/tumor. To better understand these interactions, the McBride Lab is defining the immunoglobulin repertoire of tumor-infiltrating B cells in patients who do and do not respond to immunotherapy in melanoma, renal cell carcinoma, sarcoma, and non-small cell lung cancer.
The unique pipeline developed in the lab for recombinant antibody production has been adapted, as outlined below, for studying human patient samples. In this scheme, human tumor tissue samples are enzymatically digested to release single cells. The cells are sorted and analyzed to identify and isolate B cells recognizing a single antigen through a combination of fluorescence-activated cell sorting (FACS), single cell RNA-seq, and amplification of expressed immunoglobulin genes using RT-PCR. The amplified genes are sequenced and assessed for parameters including mutation profile. The genes are then cloned into expression vectors allowing for recombinant antibody production and testing for a number of applications including immunoblotting, immunohistochemistry and enzyme-linked immunosorbent assay (ELISA). Overall, this methodology can be used to identify antibodies that recognize tumors, the specific antigens the antibodies recognize, and characterize those antibodies that recognize tumor cell surface antigens. This opens the possibility of identifying and recombinantly producing antibodies with the potential to enhance ICB efficacy in patients who lack a robust response to conventional ICB therapy. (Image created by M. Zelazowska with BioRender.com.)
Projects within this research theme include:
• Defining the immunoglobulin repertoire of tumor-infiltrating B cells in patients undergoing ICB therapy
• Characterizing B cells and the immunoglobulin repertoire in melanoma, head and neck cancer, non-small lung cancer, sarcoma, and ovarian cancer
• Working with immunologists and other collaborators toward the goal of potentiating ICB therapy by evaluating the tumor-recognizing capacity and anti-tumor efficacy of the antibodies identified through the lab's pipeline
Related Publications:
B cells and tertiary lymphoid structures promote immunotherapy response. Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, Yizhak K, Sade-Feldman M, Blando J, Han G, Gopalakrishnan V, Xi Y, Zhao H, Amaria RN, Tawbi HA, Cogdill AP, Liu W, LeBleu VS, Kugeratski FG, Patel S, Davies MA, Hwu P, Lee JE, Gershenwald JE, Lucci A, Arora R, Woodman S, Keung EZ, Gaudreau PO, Reuben A, Spencer CN, Burton EM, Haydu LE, Lazar AJ, Zapassodi R, Hudgens CW, Ledesma DA, Ong S, Bailey M, Warren S, Rao D, Krijgsman O, Rozeman EA, Peeper D, Blank CU, Schumacher TN, Butterfield LH, Zelazowska MA, McBride KM, Kalluri R, Allison J, Petitprez F, Fridman WH, Sautès-Fridman C, Hacohen N, Rezvani K, Sharma P, Tetzlaff MT, Wang L, Wargo JA. Nature. 2020 Jan;577(7791):549-555. PMID: 31942075
Mechanisms of Antibody Diversification via DNA Damage and Downstream Repair
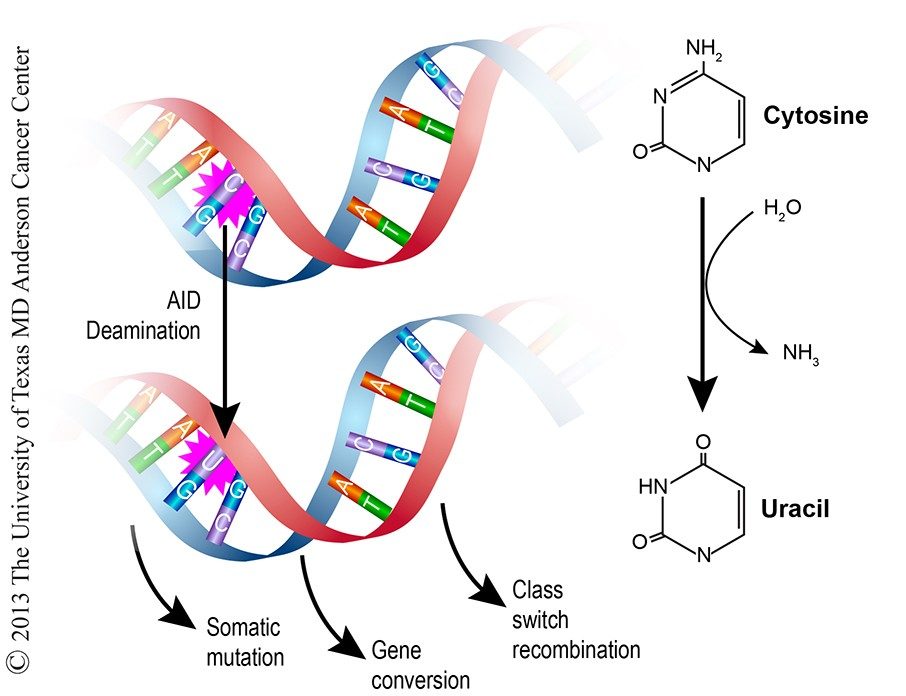
Mechanisms of Antibody Diversification via DNA Damage and Downstream Repair
The McBride Lab is defining those mechanisms that contribute to antibody diversification with an emphasis on downstream repair mechanisms. When activated, B cells migrate to germinal centers (GC), sites of B cell proliferation, immunoglobulin gene selection, maturation, and death. In the GC, B cells undergo SHM which introduces random DNA mutations into the Ig variable gene. In contrast, CSR rarely takes place in GCs, and is a deletional recombination event in which one heavy chain constant region exon is replaced by another, resulting in isotype switching (e.g. IgG, IgA, IgM) while the variable region is retained, thus also retaining antigen specificity. Both SHM and CSR are initiated and/or catalyzed by activation-induced cytidine deaminase (AID), an enzyme that introduces uracil lesions in single-stranded DNA at the Ig loci. The presence of uracil in DNA is mutagenic, compromises genomic stability, and must be repaired. Uracil lesions are commonly removed by uracil-DNA glycosylases (UNG) via base excision repair resulting in single-nucleotide mutations. Downstream repair of the DNA double-strand breaks created during CSR utilizes non-homologous end-joining mechanisms. Proper repair is needed for generating antibody diversity, maintaining genomic stability, and preventing oncogenic chromosomal translocations (e.g. IgH/MYC).
Projects within this research theme include:
• Identification and analysis of the immunoglobulin repertoire in disease states and other conditions
• Uncovering how mutation-prone versus high-fidelity repair is governed in B cells
• Defining the role of AID phosphorylation in regulating downstream DNA repair and in restricting genomic AID targets
• Characterizing the role of DNA polymerase theta and theta-mediated end-joining in antibody diversification
Related Publications:
• Targeting mutagenesis in B cells: Phosphorylation goes beyond AID association. Mu Y, McBride KM. Mol Cell Oncol. 2018 Sep 5;5(5):e1432259. PMID: 30263937
• Phosphorylation promotes activation-induced cytidine deaminase activity at the Myc oncogene. Mu Y, Zelazowska MA, McBride KM. J Exp Med. 2017 Dec 4;214(12):3543-3552. PMID: 29122947
• Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. Yousefzadeh MJ, Wyatt DW, Takata K, Mu Y, Hensley SC, Tomida J, Bylund GO, Doublié S, Johansson E, Ramsden DA, McBride KM, Wood RD. PLoS Genet. 2014 Oct 2;10(10):e1004654. PMID: 25275444
B Cells, Oncogenic Viruses, and Host-Pathogen Interactions
Viral impact on B cell development
B cells, Oncogenic Viruses, and Host-Pathogen Interactions
McBride lab members work at the intersection of virology, oncology, and immunology to discover how members of the gammaherpesvirus (GHV) family subvert B cell function.
Gammaherpesviruses, such as Epstein-Barr virus (EBV) and Kaposi sarcoma herpesvirus (KS), establish life-long latent infections that have been linked to the development of lymphomas and other cancers.
Murine herpesvirus 68 (MHV68) infection in mice drives both B
cell proliferation and lymphoma and is as a common model for
studying GHV infection and pathogenesis. It is used by the McBride
lab to identify genes that control GHV pathogenesis and determine
how GHV infection subverts host immune responses.
Upon infection, GHVs lead B lymphocytes to proliferate and diversify their Ig genes while escaping normal immune system controls. After primary infection, both EBV and MHV68 undergo latent expansion in B cells located in germinal centers (GCs) and maintain a pool of virus in isotype class-switched memory B cells. The latent virus will reactive from time-to-time and in some individuals will be triggered to become oncogenic.
The lab is defining the role of uracil DNA glycosylases (UNGs) in GHV pathogenesis and B cell development as well as the roles of host (endogenous) and viral UNG (vUNG) in establishing viral latency/chronic infection. They have discovered that the vUNG of MHV68 alters somatic hypermutation. By using single-cell and GC cell population analysis, they showed that GHV-infected germinal center B cells express a distinct and abnormal immunoglobulin repertoire. Their evidence indicates that the virus alters the immunoglobulin repertoire of the host by subverting host B cell selection and immunoglobulin evolution.

Projects within this research theme include:
• Characterization of the B cell subtype inhabited by gammaherpesviruses in vivo
• Analyzing mechanisms of gammaherpesvirus subversion of host immunoglobulin repertoire
• Identification and characterization of viral genes involved in host immune subversion
• Defining the differences in how gammaherpesvirus vUNG and host UNG aid DNA repair by biochemically characterizing vUNG
Related Publications:
• Multifaceted roles for STAT3 in gammaherpesvirus latency revealed through in vivo B cell knockout models. Hogan CH, Owens SM, Reynoso GV, Liao Y, Meyer TJ, Zelazowska MA, Liu B, Li X, Grosskopf AK, Khairallah C, Kirillov V, Reich NC, Sheridan BS, McBride KM, Gewurz BE, Hickman HD, Forrest JC, Krug LT. mBio. 2024 Feb 14;15(2):e0299823. PMID: 38170993
• Uracil-DNA glycosylase of murine gammaherpesvirus 68 binds cognate viral replication factors independently of its catalytic residues. Smith KR, Paul S, Dong Q, Anannya O, Oldenburg DG, Forrest JC, McBride KM, Krug LT. mSphere. 2023 Oct 24;8(5):e0027823. PMID: 37747202
• Divergent structures of Mammalian and gammaherpesvirus uracil DNA glycosylases confer distinct DNA binding and substrate activity. Mu Y, Zelazowska MA, Chen Z, Plummer JB, Dong Q, Krug LT, McBride KM. DNA Repair (Amst). 2023 Aug;128:103515. PMID: 37315375
• Dangerous Liaisons: Gammaherpesvirus Subversion of the Immunoglobulin Repertoire. Zelazowska MA, McBride K, Krug LT. Viruses. 2020 Jul 23;12(8):788. PMID: 32717815
• Gammaherpesvirus-infected Germinal Center Cells Express a Distinct Immunoglobulin Repertoire. Zelazowska MA, Dong Q, Plummer JB, Zhong Y, Liu B, Krug LT, McBride KM. Life Sci Alliance. 2020 Feb 6;3(3):e201900526. PMID: 32029571
• Combinatorial Loss of the Enzymatic Activities of Viral Uracil-DNA Glycosylase and Viral dUTPase Impairs Murine Gammaherpesvirus Pathogenesis and Leads to Increased Recombination-Based Deletion in the Viral Genome. Dong Q, Smith KR, Oldenburg DG, Shapiro M, Schutt WR, Malik L, Plummer JB, Mu Y, MacCarthy T, White DW, McBride KM, Krug LT. mBio. 2018 Oct 30;9(5):e01831-18. PMID: 30377280
• Absence of the uracil DNA glycosylase of murine gammaherpesvirus 68 impairs replication and delays the establishment of latency in vivo. Minkah N, Macaluso M, Oldenburg DG, Paden CR, White DW, McBride KM, Krug LT. J Virol. 2015 Mar;89(6):3366-79. PMID: 25589640
Recent Funding
The Mark Foundation For Cancer Research: Harnessing tertiary lymphoid structures for improved immunotherapeutic strategies in cancer patients (02/01/2022-1/31/2025)
Cancer Prevention Research Institute of Texas (CPRIT) High Impact High Risk Award, Project number RP200574: Targeting B cells to enhance responses to immune checkpoint blockade (08/31/2020-08/30/2022)
Cancer Prevention Research Institute of Texas (CPRIT) Core Facility Support Award, Project number RP190507: Recombinant Antibody Production Core (RAPC) 08/31/2019-08/30/2024
Leukemia & Lymphoma Society Specialized Center of Research Program Award, Project number SCOR-12206-17: Role of WWOX Loss of Function in High-Risk Multiple Myeloma 10/01/2017-09/30/2022
NIH/NIAID Research Project Grant Program Award, Project number R01AI121403: Modeling ICF syndrome in mice: Role of Zbtb24 in DNA methylation and antibody production (09/26/2016-08/31/2022)
NIH/NIAID Research Project Grant Program Award, Project number R01AI125397: Uracil DNA Glycosylases in Gammaherpesvirus Pathogenesis and B Cell Development (06/01/2016-05/31/2022)
Recombinant Antibody Production & Creation Platform

Antibodies are a central tool in biomedical science: they allow researchers to identify, purify, and track molecular targets; however, it is not always possible to obtain a supply of replenishable, consistently reliable, and molecularly characterized antibodies. Our process uses single-cell cloning technology to purify cells expressing antigen-specific antibodies via fluorescence-activated cell sorting directly from immunized mice. Immunoglobulin genes are amplified from single cells, cloned into expression vectors, and expressed in vitro to produce antibodies.
Most contemporary antibody production methodologies are slow, expensive, low-throughput, and cannot guarantee the resulting antibodies will be suited for their intended purpose. Many commercial antibodies have not been defined at the molecular level and can be of poor quality which wastes precious research resources and contributes to the so-called “reproducibility crisis” in biomedical research.
Pilot project funding supported establishing and optimizing the protocols used to clone immunoglobulin genes and produce recombinant antibodies, leading to refined methods for isolating specific immunoglobulin genes for future antibody production, and the formation of a Recombinant Antibody Production Service that evolved into the multi-service core Recombinant Antibody Production Core, under CPRIT grant RP190507. Now that the grant has ended (August 2024), the core is transitioning into a Recombinant Antibody Production and Creation Platform.
Advantages of recombinant monoclonal antibody production technology over traditional hybridoma production include:
- Potential for limitless supply with consistent quality
- Antibody reconfiguration or fusion to epitope tags, fluorescent protein or HRP
- Better suited to producing modification-specific antibodies, as screening can potentially occur prior to cloning and production
The Platform
The Recombinant Antibody Production and Creation Platform (RAPCP) provides custom recombinant monoclonal antibodies using a rapid, high-throughput, and cost-effective platform. The McBride Lab developed a propriety method for isolating single-antigen-specific plasma cells following inoculation into mice. Cells are sorted, the immunoglobulin genes are amplified and sequenced, and the sequences are analyzed. Curated antibody-encoding genes are cloned into expression vectors for recombinant antibody production and purification. The antibodies can then be tested in collaboration with the end-user, using a number of assays based on user priorities. This process results in sequence-defined, recombinant antibodies in as few as 30-90 days. McBride lab members will consult and collaborate with you on selecting and targeting your antigen, while also aiding in assay development to vet your antibodies with respect to antigen targeting, antibody specificity, and antibody affinity.
Instrumentation
The process to make recombinant antibodies uses state-of-the art equipment to generate sequence-defined, recombinant antibodies in as little as one month. They are able to rapidly isolate individual B cells using a BD FACS ARIA Fusion and ready the single cells for RT PCR using an epMotion liquid handler. PCR is accomplished rapidly using an Applied Biosystems Veriti 384-well thermocycler coupled with an Agilent Zag DNA Analyzer for parallel capillary electrophoresis for amplicon analysis. Selected, purified DNA samples are sequenced and selected before matched heavy and light chain genes are cloned into expression vectors and co-electroporated into modified CHO cells using a BTX HT-100 high-throughput electroporation system. Antibody-expressing cells are grown in a Kuhner CO2 shaker-incubator and the expressed antibodies are harvested and purified via protein-A binding in automated 96-well format.
Custom antibody creation and patient antibody cloning
Custom antibody creation:
The Recombinant Antibody Production and Creation Platform’s unique workflow performs both antigen screening and sequence analysis in the first steps of the process. Screening and cloning antibodies takes place up-front, not post-antibody production. In contrast to hybridoma technology, this process does not require high antibody titers. Fully recombinant antibodies overcome many obstacles of classical methods such as antibody reproducibility, ill-defined or non-defined antibody sequence, and poor antibody characterization. The techniques used by the platform are especially well-suited for generating antibodies that recognize challenging targets, such as antigens with single amino acid polymorphisms or post-translational modifications. The antibodies can be cloned into desired configurations such as single-chain or bispecific antibodies.
• Recombinant antibody project collaborations (feasibility, antigen design)
-Fully human antibodies using immunized Alloy Therapeutics ATX-GK mice
-Cellular production and purification (surface or intracellular proteins) of antigens
-Antibodies recognizing post-translational modifications using peptide antigens
• Recombinant antibody production, purification, and high-throughput characterization
-Immunoglobulin sequencing
-Immunoglobulin cloning
-Antibody validation assays (cell surface binding, antibody-dependent cellular toxicity, internalization assays, etc.)
• Recombinant antibody engineering
-Fluorochrome coupling, or fusing epitope, or affinity tags
-Single-chain or bispecific antibodies
-Different species Fc or isotypes.
• Conversion of existing hybridoma cell lines to recombinant antibody production stocks
Patient antibody cloning:
• Antibody profiling of human tumors (e.g. melanoma, sarcoma, non-small cell lung cancer, ovarian cancer)
-Profiling the immunoglobulin repertoire of tumor-associated B cells from tumor samples or single-cell seq data
-Recapitulating antibodies expressed by tumor-associated B cells
-Defining B cell clonal expansion, evolution, and selection
-Comparing circulating B cell populations and tumor-associated B cell populations
Recombinant Antibody Projects
Overview - how recombinant antibodies are produced
The McBride lab's overall process includes several platforms used for isolation of single B cells, the recovery of immunoglobulin sequences by immunoglobulin profiling and sequence analysis, cloning and expressing the immunoglobulin genes, and purifying and testing the resulting recombinant antibodies.
Currently, they can perform immunoglobulin recovery on B cells isolated from patient tumor samples, peripheral blood mononuclear cells (PBMCs), and mouse spleens. Immunoglobulin profiling uses 5’ single-cell RNA-seq or B-cell receptor-seq. Sequences are analyzed using one or more methods, and the selected immunoglobulin-encoding sequences are cloned into expression vectors. The cloned genes are expressed in cells to recombinantly produce antibodies. The antibodies are then purified and tested in one or more assays.
Overview - production of recombinant antibodies against viral uracil-DNA glycoslylase (vUNG)
In this example, the lab developed antibodies against gammaherpesvirus uracil DNA glycosylase that recognize the viral protein (vUNG) but not the related mouse protein. Their approach identifies those cells that recognize the antigen of interest early in the process, through a combination of antigen binding and fluorescence-activated cell sorting.
Capture of antigen-specific plasma cells (vUNG)
In this example, plasma cells expressing antibodies specifically recognizing a recombinant gammaherpes virus antigen (vUNG) inoculated into mice were separated from those displaying non-specific binding.
Defining the immunoglobulin repertoire (vUNG)
Expressing and testing the recombinant antibodies (vUNG)
Finally, the immunoglobulin genes encoding the antibody heavy and light chains are cloned into expression vectors and transfected into cells that will secrete the antibodies.
The secreted antibodies are purified using a high-throughput, automated protein-A purification system. Antibody integrity is checked by SDS-PAGE, and the antibodies are characterized in a downstream assay, in this case an ELISA.
The ELISA data, graphed below, illustrate the specificity of the recombinant anti-vUNG antibodies against the target v-UNG protein (left) compared to the related, non-target murine UNG (right).
Recombinant antibodies recognizing post-translationally modified protein (AID phospho-serine 38)
In this example project, the McBride group generated antibodies that recognized a post-translationally modified protein (phosphorylated activation-induced deaminase) but not the unmodified protein.
The workflow diagram illustrates the process of producing this antibody from the inoculation of mice to the testing of the recombinantly expressed antibodies by immunoblotting.
Molecularly engineered recombinant antibodies
Give Now
Research Areas
Find out about the four types of research taking place at UT MD Anderson.
