
Bartholomew Laboratory
Blaine Bartholomew, Ph.D.
Principal Investigator
- Departments, Labs and Institutes
- Labs
- Bartholomew Laboratory
Area of Research
- Epigenetics
- Chromatin Remodelers
- Biochemistry
- Histone Modification
- Chromatin
The Bartholomew laboratory studies ATP-dependent chromatin remodelers, a class of epigenetic factors that regulate chromatin structure and function in an ATP-dependent manner. Gaining a better understanding of ATP-dependent remodeling complexes is vital for improving human health as mutations in these complexes can drive both cancer and other developmental disorders. The lab is defining how chromatin remodeling complexes regulate gene expression, DNA repair, and nuclear architecture and is identifying the functional differences between family members, focusing on molecular mechanisms underlying the normal and altered states of these complexes. A key tenet of the lab is to maintain a collaborative and supportive environment, committed to both learning and professional excellence.
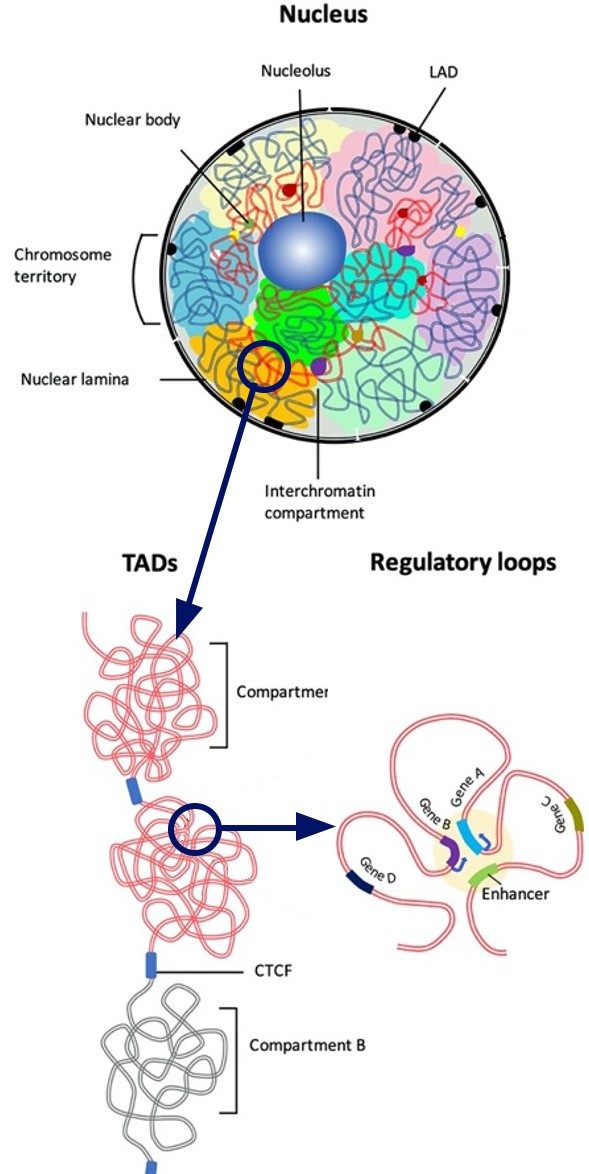
Nuclear Organization, Cell Differentiation & Chromatin Remodeling
The mammalian SWI/SNF complex, whose functional roles are just beginning to be deciphered, is a master regulator in development. Mutations in the SWI/SNF complex are the driving cause for many human diseases including cancer.
Although SWI/SNF is highly enriched at transcriptional enhancers and its basic chromatin remodeling activities have been studied for over 30 years, there is little known about how it regulates enhancer activity and enhancer-promoter interactions.
By targeting a DNA/RNA binding module called the AT-hook, which resides in the catalytic subunit of the megadalton SWI/SNF complex, the Bartholomew lab has uncovered a role for SWI/SNF in de novo enhancer activation and nuclear architecture.
The model system the lab uses to study the role of SWI/SNF in both enhancer activation and regulation of nuclear architecture is stem cells during their transition from a naïve pluripotent state to a state putting them squarely on a path toward cell lineage priming and cell fate determination. During this transition, there is both significant restructuring of the nuclear architecture and transcriptional rewiring, including de novo enhancer activation, that makes this an exquisitely ideal system for studying the SWI/SNF complex.

Chromatin Dynamics, Transcription Regulation, DNA Replication & Repair
The INO80 ATP-dependent chromatin remodeler operates at promoters, telomeres and centromeres; and is involved in transcription regulation, 3-D organization of the genome, replication, DNA repair, and heterochromatin/centromere formation. Several lines of evidence show that INO80 promotes nucleosome disassembly and exchange of H2A.Z-H2B dimers, but current biochemical data has not yet revealed how INO80 disrupts chromatin. The Bartholomew lab is defining how INO80 promotes loss of canonical H3 at centromeres, promotes the exchange of H2A.Z at DNA double strand breaks, pericentric and other regions, and increases enhancer accessibility in several cancers by more fully understanding the mechanism of INO80 chromatin remodeling. The lab is also working to uncover how DNA sequence contributes to large differences in the efficiency and possibly the ultimate outcomes of INO80 remodeling.
Join our ranks! Many of those who have trained in the Bartholomew Lab have gone on to exceptional postdoc positions, become successful faculty members, principal investigators, and leaders in the biotech and pharmaceutical industries.
We are leaders in defining:
Nuclear Organization | Chromatin Remodeling & Dynamics | Cell Differentiation | Transcription Regulation | DNA Replication & Repair
Give Now
Research Areas
Find out about the four types of research taking place at UT MD Anderson.
