Epigenomics Profiling Core
- Research Resources
- Core Facilities and Services
- Epigenomics Profiling Core
Overview
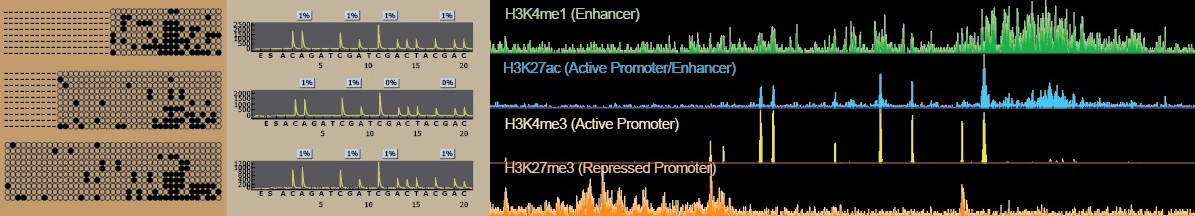
The Epigenomics Profiling Core (EpiCore) is an institutional resource for epigenomics research that brings scientific expertise, advanced assays and specialized equipment not readily available to UT MD Anderson researchers. We offer cost-effective, full-service protocols to investigate DNA methylation (Pyrosequencing Methylation Analysis, RRBS, cfMeDIP-seq, WGBS), chromatin accessibility (ATAC-Seq) and DNA-protein interactions (ChIP-Seq and CUT&RUN). Our overall goal is to facilitate researchers’ efforts to understand epigenetic mechanisms that alter the chromatin state that regulate the transcription output in normal and disease conditions. Learn more about the Epigenomics Profiling Core.
Getting Started
Each project starts with a consultation with core directors to design experimental option(s). Please contact Marcos Estecio, Ph.D., (mestecio@mdanderson.org) for DNA methylation analysis services and Abhinav Jain, Ph.D., (ajain@mdanderson.org) for chromatin analysis services (ChIP-seq, CUT&RUN and ATAC-seq).
For additional scheduling, please use iLab.
Impact
We offer a one-stop unique resource for expertise, methods and services to assess the epigenome for basic and clinical applications, such as determining effectiveness of epigenetic therapies. The core directors work closely with the investigators to adapt protocols to match the needs of specific biological samples, data interpretation, manuscript and grant preparation.
New Services:
We are now offering the following services:
- HiC/HiChIP assays for 3D chromatin organization
- cfChIP-Seq of cell-free DNA from liquid biopsies to sequence DNA fragments from nucleosomes carrying specific chromatin marks
- Bioinformatics Analysis of ChIP-Seq, ATAC-Seq and RRBS libraries
- cfMeDIP (Circulating Cell-free Methylated DNA Immunoprecipitation)
- CUT&RUN (Cleavage Under Targets and Release Using Nuclease)
Services Available Soon
cfChIP-Seq of cell-free DNA from liquid biopsies
Chromatin Immunoprecipitation and sequencing of cell-free nucleosomes from human plasma (cfChIP-seq) to sequence DNA fragments from nucleosomes carrying specific chromatin marks
Direct DNA and RNA modifications profiling by Nanopore
The EpiCore has recently acquired a GridION MK1 Sequencing Instrument (Oxford Nanopore Technologies) and will soon offer direct detection of base modifications in DNA and RNA.
Contact
DNA Methylation Analysis Services
Marcos R. Estecio, Ph.D.
Co-director, Epigenomics Profiling Core
Office phone: 713-792-9108
Email: mestecio@mdanderson.org
Chromatin Analysis Services
(cfChIP-seq, ChIP-seq, CUT&RUN and ATAC-seq)
Abhinav Jain, Ph.D.
Co-director, Epigenomics Profiling Core
Office phone: 713-745-2640
Email: ajain@mdanderson.org
Location:
UT MD Anderson
Basic Sciences Research Building - S9.8414
6767 Bertner Ave.
Houston, TX 77030
Give Now
Research Areas
Find out about the four types of research taking place at UT MD Anderson.
